 Author
Author  Correspondence author
Correspondence author
International Journal of Clinical Case Reports, 2024, Vol. 14, No. 4
Received: 01 Jun., 2024 Accepted: 09 Jul., 2024 Published: 03 Aug., 2024
The HOXB13 gene has emerged as a key factor in prostate cancer susceptibility, particularly its most notable variant, G84E, which is associated with a significantly increased risk of prostate cancer, especially in individuals with a family history or early-onset disease. This report explores the genetic mechanisms of HOXB13 variants, their role in regulating androgen receptor (AR) signaling, and their impact on prostate cancer aggressiveness and treatment resistance. The clinical applications of HOXB13 variants in early diagnosis, risk stratification, and personalized treatment strategies are also discussed. While HOXB13 shows great potential as a biomarker, challenges remain in translating these findings into widespread clinical applications, particularly in non-European populations. Future research must focus on large-scale population studies and the development of therapeutic strategies targeting HOXB13-driven pathways to enhance the management of prostate cancer.
1 Introduction
Prostate cancer (PrCa) is a multifactorial disease influenced by both genetic and environmental factors. Inherited genetic predispositions contribute significantly to its onset, especially among men with a family history of prostate cancer. Variants in several genes have been associated with increased prostate cancer risk, with HOXB13 playing a particularly critical role. It has been established that men with certain germline mutations in key susceptibility genes, such as BRCA2, ATM, and HOXB13, have a heightened risk of developing prostate cancer. Studies highlight that prostate cancer risk is increased particularly in families with early-onset and hereditary cases of the disease (Kote-Jarai et al., 2015).
The HOXB13 gene encodes a transcription factor crucial for prostate development. HOXB13 regulates androgen receptor signaling, which is essential for normal prostate cell proliferation and differentiation. The most notable variant, G84E, has been strongly associated with familial and early-onset prostate cancer (Kurihara et al., 2022). This variant is rare but confers a significantly higher risk of developing the disease, particularly in European populations. Research indicates that G84E homozygotes or heterozygotes have a markedly increased risk for prostate cancer, with some studies estimating an 8-fold higher risk in affected individuals (Sipeky et al., 2018). Furthermore, HOXB13 variants are involved in more aggressive forms of prostate cancer, as synergistic interactions with other genes such as CIP2A exacerbate the disease's severity (Dupont et al., 2021).
This report provides a comprehensive genetic analysis of HOXB13 variants and their clinical significance in the diagnosis and treatment of prostate cancer. By exploring the mechanisms through which HOXB13 mutations contribute to the onset and aggressiveness of prostate cancer, it aims to deepen the understanding of the roles of these genetic variants. Understanding these interactions can assist clinicians in better stratifying patients for screening and treatment based on their genetic risk, thereby improving personalized prostate cancer management strategies. The report also reviews the potential of HOXB13 as a biomarker for early detection and prognosis in high-risk individuals.
2 Genetic Analysis of HOXB13 Gene Variants
2.1 Overview of the structure and function of the HOXB13 gene
The HOXB13 gene, located on chromosome 17q21, is a member of the Homeobox gene family, which encodes transcription factors responsible for regulating the development of various tissues. In particular, HOXB13 plays a key role in the differentiation and development of the prostate gland, influencing androgen receptor (AR) signaling (Tilki et al., 2016). The G84E mutation in HOXB13 has been identified as a significant genetic variant, conferring susceptibility to prostate cancer. The gene comprises two main exons that encode a protein critical for maintaining normal prostate function. Mutations in the HOXB13 gene disrupt these processes, contributing to cancer development by promoting uncontrolled cell proliferation (Sipeky et al., 2018).
2.2 Types of HOXB13 gene mutations and their distribution in prostate cancer
The G84E variant (rs138213197) is the most studied mutation in the HOXB13 gene, predominantly associated with hereditary prostate cancer. This mutation is characterized by a guanine-to-adenine substitution, leading to the replacement of glycine with glutamic acid at position 84. It has been linked to increased prostate cancer risk, especially in men of European descent (Boyle et al., 2020). Other rare variants, such as G132E and F127C, have also been reported in specific populations, such as in Japanese men with familial prostate cancer (Kurihara et al., 2022). These mutations vary significantly across different geographic and ethnic groups, affecting risk levels and disease severity (Lotan et al., 2017).
2.3 Studies on the association between HOXB13 gene variants and the risk of prostate cancer
Multiple studies have established a strong link between HOXB13 mutations and an increased risk of prostate cancer. The G84E variant, in particular, has been identified as a moderate-penetrance mutation that significantly increases the risk of developing prostate cancer, especially in men with a family history of the disease. Research by Dupont et al. (2021) found that carriers of the G84E mutation have up to an 8-fold increased risk of developing prostate cancer compared to non-carriers, especially those diagnosed with early-onset or aggressive forms of the disease. Additional studies have shown that the mutation is prevalent in approximately 1.5% of prostate cancer cases but is absent or rare in non-cancer populations (Kote-Jarai et al., 2015). Furthermore, studies conducted in diverse populations, including Finnish and British men, emphasize the mutation’s role in familial prostate cancer predisposition (Sipeky et al., 2018).
2.4 Influence of ethnicity, family history, and other genetic factors on HOXB13 variants
The distribution and impact of HOXB13 variants, particularly the G84E mutation, show significant variation based on ethnicity and familial predisposition (Wang, 2024). Studies have found that G84E is predominantly prevalent among men of European descent, where it is strongly associated with familial prostate cancer. In contrast, other mutations such as G132E and F127C have been identified in Japanese populations, with varying degrees of risk implication (Kurihara et al., 2022). The risk of developing prostate cancer for HOXB13 mutation carriers also increases significantly in those with a positive family history of the disease. Additionally, HOXB13 mutations can interact synergistically with other genetic variants, such as CIP2A mutations, to further exacerbate disease risk, particularly in aggressive and early-onset cases (Sipeky et al., 2018).
3 Molecular Mechanisms of HOXB13 Gene Variants
3.1 Impact of HOXB13 gene variants on gene expression and regulatory pathways
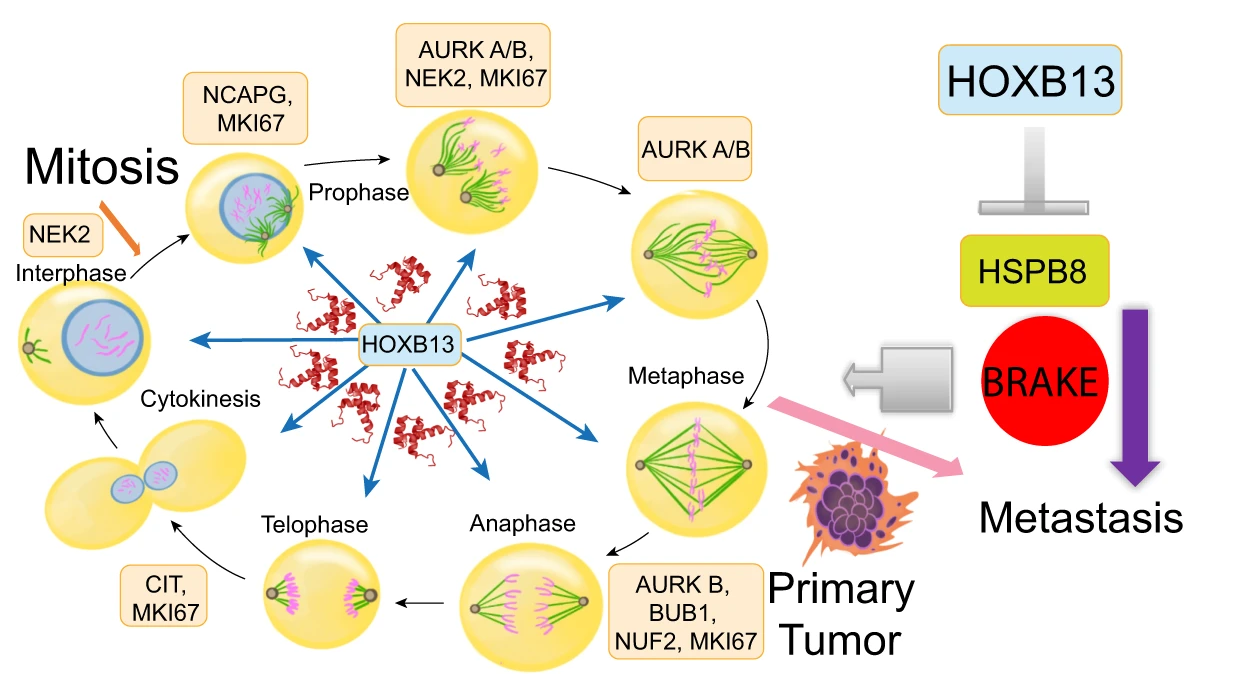
The HOXB13 gene plays a key role in regulating gene expression through its function as a transcription factor. Variants in HOXB13, particularly the G84E mutation, have been shown to alter gene expression patterns in prostate cancer cells, influencing pathways involved in cell proliferation and metastasis (Heise et al., 2019). HOXB13 binds to androgen receptor (AR)-regulated elements and modulates the activity of genes involved in prostate development and tumor progression. Studies suggest that HOXB13 regulates mitotic protein-kinase interaction networks in metastatic prostate cancers, driving aggressive cancer phenotypes by activating a subset of genes essential for cancer cell proliferation and migration (Figure 1) (Yao et al., 2019).
|
Figure 1 A schematic showing key mitotic kinases over-expressed in metastatic PCs and regulated by HOXB13. HSPB8 functions as a restraint against PC metastasis (Adopted from Yao et al., 2019) |
3.2 Interaction between HOXB13 and androgen receptor signaling pathways
HOXB13 interacts closely with the androgen receptor (AR), which is critical for prostate cancer progression. In castration-resistant prostate cancer (CRPC), HOXB13 acts as a co-regulator of AR and its splice variant AR-V7, facilitating ligand-independent AR-driven transcriptional activity (Song et al., 2024). This interaction has been linked to the development of resistance to androgen deprivation therapy (ADT) and is associated with poor prognosis in prostate cancer patients. HOXB13 enhances the expression of AR target genes involved in cell survival and proliferation, thus promoting tumor growth even in the absence of androgens (Coutinho et al., 2016; Chen et al., 2018). Additionally, HOXB13 cooperates with other co-regulators like FOXA1 and GATA2 to activate AR signaling, which contributes to the metastatic potential of prostate cancer (Hankey et al., 2020).
3.3 Relationship between HOXB13 gene variants and prostate cancer metastasis and progression
HOXB13 expression is significantly associated with increased prostate cancer metastasis and disease progression. High levels of HOXB13 have been correlated with advanced tumor stages and poor clinical outcomes following radical prostatectomy. In particular, HOXB13 promotes metastasis through the regulation of bone metastasis-related genes, such as those involved in cytokine signaling and integrin-mediated adhesion, which enhance the ability of cancer cells to invade bone tissue. Studies have shown that the HOXB13-HOXA11-AS axis modulates bone metastasis by regulating critical genes like CCL2/CCR2 and IBSP, which are involved in cell invasion and the formation of metastatic lesions (Misawa et al., 2021). Moreover, HOXB13 interacts with AR-V7 to drive prostate cancer cell migration and metastasis, particularly in castration-resistant cases, by maintaining chromatin accessibility at AR-binding sites that promote aggressive cancer phenotypes (Sharp et al., 2018).
4 Clinical Applications and Predictive Value
4.1 HOXB13 gene variants as early diagnostic markers for prostate cancer
The HOXB13 G84E mutation has emerged as a promising biomarker for early detection of prostate cancer, especially in men with a family history of the disease. Several studies have demonstrated that men carrying the G84E variant have a significantly increased risk of developing prostate cancer, particularly at an early age. Population-based studies have established that G84E can stratify high-risk individuals for earlier screening and more intensive monitoring (Kote-Jarai et al., 2015). This variant's role in early diagnosis is especially significant in populations of European descent, where it has been linked to a greater likelihood of hereditary prostate cancer.
4.2 Significance of HOXB13 gene variants in prostate cancer risk assessment
HOXB13 gene mutations, particularly G84E, are associated with a marked increase in prostate cancer risk. The G84E mutation contributes to a 3-fold higher risk of developing the disease compared to non-carriers (Maia et al., 2015). Studies show that this variant is particularly valuable for assessing risk in individuals with a family history of prostate cancer, where the odds ratio can be even higher (Figure 2) (Dupont et al., 2021). Moreover, risk assessment models incorporating HOXB13 variants alongside other genetic markers such as BRCA2 and ATM have proven effective in identifying individuals at higher risk for aggressive prostate cancer (Sipeky et al., 2018).
|
Figure 2 Association of genetic variants of the HOXB locus with prostate cancer in combined ICPCG, NFPCS and PLCO subjects (Adopted from Dupont et al., 2021) |
4.3 Potential application of HOXB13 gene variants in personalized treatment
HOXB13 mutations have potential applications in guiding personalized treatment strategies for prostate cancer. As HOXB13 interacts with androgen receptor (AR) signaling, the presence of specific variants may indicate susceptibility to certain therapies, such as androgen deprivation therapy (ADT) or AR-targeted treatments (Park et al., 2019). Recent studies have highlighted that the expression of HOXB13 may modulate the efficacy of therapies targeting AR, thus influencing treatment outcomes in advanced and castration-resistant prostate cancer (Patel et al., 2023). Understanding these variants' role in treatment response allows for the development of more tailored therapies that address the molecular mechanisms driving individual tumors (Johng et al., 2016).
4.4 Role in predicting prostate cancer recurrence and prognosis
HOXB13 overexpression has been shown to predict early prostate-specific antigen (PSA) recurrence following radical prostatectomy. Patients exhibiting high levels of HOXB13 expression have poorer outcomes, as this variant is linked to more aggressive disease and higher Gleason scores. HOXB13 variants, in combination with AR and PSA levels, provide strong prognostic information about recurrence and overall survival after treatment (Zabalza et al., 2015). Further studies have demonstrated that HOXB13 can be used as a marker for identifying patients at higher risk of biochemical recurrence, particularly when combined with other genetic markers like CIP2A (Sipeky et al., 2018).
5 HOXB13 Gene Variants as Therapeutic Targets
5.1 Targeted therapy strategies based on HOXB13 variants
HOXB13 is a critical player in androgen receptor (AR) signaling, and therapies targeting HOXB13's interaction with AR and its variants are showing promise. HOXB13 is essential for the regulation of AR splice variant-7 (AR-V7), a key driver of castration-resistant prostate cancer (CRPC). Studies indicate that silencing HOXB13 in CRPC models reduces AR-V7 oncogenic functions, providing a potential therapeutic target for AR-V7-driven tumors (Chen et al., 2018). Another promising strategy involves inhibiting acetylated HOXB13, which primes CRPC for AR antagonism by regulating super-enhancer genes that drive cancer progression (Nguyen et al., 2022). These findings suggest that targeting HOXB13 could be an effective approach in personalized treatment for prostate cancer.
5.2 Potential impact of HOXB13 gene variants on immunotherapy response
The impact of HOXB13 gene variants on immunotherapy responses is an emerging area of research. Although the interaction between HOXB13 and immune checkpoint inhibitors has not been fully explored, its role in regulating androgen receptor-driven pathways and tumor progression may influence immune evasion mechanisms in prostate cancer. Current research focuses on how targeting HOXB13-regulated pathways might improve the efficacy of immune checkpoint blockade therapies. Further studies are needed to clarify the direct link between HOXB13 expression and immunotherapy outcomes in metastatic CRPC (Nguyen et al., 2022).
5.3 Relationship between HOXB13 gene mutations and resistance to chemotherapy or hormonal therapy
HOXB13 mutations, particularly G84E, are associated with resistance to traditional treatments, such as androgen deprivation therapy (ADT) and chemotherapy. Research has shown that HOXB13 co-regulates AR splice variants, including AR-V7, which are known to drive resistance to AR-targeting agents like enzalutamide and abiraterone (Liu et al., 2021). By promoting AR-V7 expression, HOXB13 enables tumor cells to evade hormonal therapies, making it a key factor in therapy resistance (Chen et al., 2018). Targeting HOXB13 in conjunction with AR antagonists may improve treatment responses, particularly in advanced cases of castration-resistant prostate cancer (Sipeky et al., 2018).
6 Future Research Directions and Challenges
6.1 Other potential mechanisms and functions of HOXB13 gene variants
While the role of HOXB13 mutations, particularly G84E, in prostate cancer is well-documented, ongoing research is needed to uncover additional mechanisms and functions of HOXB13 in prostate cancer progression. Recent studies suggest that HOXB13 influences key oncogenic pathways by interacting with chromatin remodelers and transcription factors like androgen receptors (AR) (Nerlakanti et al., 2018; Lu et al., 2021). Beyond AR, HOXB13 also impacts genes involved in mitotic regulation and cancer cell migration, pointing to its broader role in cancer metastasis. The identification of downstream targets, such as the mitotic kinase CIT and the tumor suppressor HSPB8, has highlighted novel pathways that HOXB13 might regulate in metastatic prostate cancer, offering opportunities for future therapeutic interventions (Yao et al., 2019).
6.2 Necessity for large-scale population studies on HOXB13 gene variants
The current data on HOXB13 gene variants, such as G84E, have predominantly focused on European populations. However, to understand the true global impact of these variants, large-scale population studies across diverse ethnic groups are essential. Such studies can reveal ethnic differences in the prevalence of HOXB13 mutations and assess their association with prostate cancer risk in non-European populations (Schnoeller et al., 2015). For example, the G132E and F127C variants have been associated with prostate cancer risk in Japanese men, illustrating the importance of expanding research to other populations (Kurihara et al., 2022). Additionally, combining genetic data from large cohorts like the UK Biobank has proven effective in identifying novel risk loci beyond HOXB13, which further emphasizes the need for large-scale, multi-ethnic studies (Emami et al., 2020).
6.3 How to apply HOXB13 gene variant research in clinical practice
The integration of HOXB13 variant research into clinical practice holds great promise for improving prostate cancer diagnosis, prognosis, and treatment. However, the challenge lies in translating genetic findings into practical tools for clinicians (Kim et al., 2020). HOXB13 variants such as G84E should be incorporated into genetic screening panels for early detection and risk assessment, particularly in individuals with a family history of prostate cancer (Weiner et al., 2020). Moreover, research into how HOXB13 interacts with other prostate cancer biomarkers, such as AR-V7, may pave the way for personalized treatment strategies (Feng et al., 2021). For instance, targeting HOXB13 in combination with androgen receptor antagonists might improve outcomes in castration-resistant prostate cancer (CRPC) (Chen et al., 2018). Future clinical trials should explore how these genetic insights can be utilized for more tailored treatment approaches.
7 Concluding Remarks
The HOXB13 gene, particularly the G84E variant, has been identified as a key genetic factor in the development and progression of prostate cancer. This variant significantly increases the risk of prostate cancer, especially in individuals with a family history of the disease or early-onset prostate cancer. HOXB13's role in regulating androgen receptor (AR) signaling, which is critical for prostate cell growth, highlights its importance in prostate cancer biology. Furthermore, its interactions with other genes, such as CIP2A, and pathways involved in cell proliferation, have been shown to predispose individuals to more aggressive forms of the disease. Variants such as G132E and F127C, identified in Japanese and other non-European populations, further demonstrate the global relevance of HOXB13 in prostate cancer risk.
The potential for HOXB13 variants, particularly G84E, as biomarkers for early diagnosis and risk stratification is promising. In clinical practice, screening for these variants could enable the identification of high-risk individuals, guiding personalized approaches to screening and treatment. However, challenges remain in applying HOXB13 variants to widespread clinical practice. The rarity of the G84E mutation limits its utility in general populations, necessitating larger-scale studies in diverse populations to identify additional variants that could be more broadly applicable. Additionally, the role of HOXB13 in resistance to therapies, such as androgen deprivation therapy (ADT) and chemotherapy, presents both opportunities for targeted treatments and challenges in overcoming therapy resistance. As research continues, refining genetic screening guidelines and developing targeted therapies based on HOXB13 variants will be essential in improving clinical outcomes for prostate cancer patients.
Acknowledgment
Thank you to the anonymous reviewers for their suggested revisions to this study.
Conflict of Interest Disclosure
The author affirms that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Boyle J., Hahn A., Kapron A., Kohlmann W., Greenberg S., Parnell T., and Cooney K., 2020, Pathogenic germline DNA repair gene and HOXB13 Mutations in men with metastatic prostate cancer, JCO Precision Oncology, 4: PO.19.00284.
https://doi.org/10.1200/po.19.00284.
PMID: 32923906 PMCID: PMC7446531
Chen Z., Wu D., Thomas-Ahner J., Lu C., Zhao P., Zhang Q., Geraghty C., Yan P., Hankey W., Sunkel B., Cheng X., Antonarakis E., and Wang Q., 2018, Diverse AR-V7 cistromes in castration-resistant prostate cancer are governed by HOXB13, Proceedings of the National Academy of Sciences, 115(26): 6810-6815.
https://doi.org/10.1073/pnas.1718811115
PMID: 29844167 PMCID: PMC6042123
Coutinho I., Day T.K., Tilley W., and Selth L., 2016, Androgen receptor signaling in castration-resistant prostate cancer: a lesson in persistence, Endocrine-related Cancer, 23(12): 179-197.
https://doi.org/10.1530/ERC-16-0422
PMID: 27799360
Dupont W., Breyer J.P., Johnson S.P., Plummer W., and Smith J.R., 2021, Prostate cancer risk variants of the HOXB genetic locus, Scientific Reports, 11: 89399.
https://doi.org/10.1038/s41598-021-89399-7
PMID: 34059701 PMCID: PMC8167119
Emami N.C., Cavazos T.B., Rashkin S., Cario C.L., Graff R.E., Tai C., Mefford J., Kachuri L., Wan E., Wong S., Aaronson D., Presti J., Habel L., Shan J., Ranatunga D., Chao C., Ghai N.R., Jorgenson E., Sakoda L., and Witte J., 2020, A large-scale association study detects novel rare variants, risk genes, functional elements, and polygenic architecture of prostate cancer susceptibility, Cancer Research, 81: 1695-1703.
https://doi.org/10.1158/0008-5472.CAN-20-2635
PMID: 33293427 PMCID: PMC8137514
Feng T., Wang J., Cheng K., Lu Q., Zhao R., Wang S., Zhang Q., Ge L., Pan J., Song G., and Wang L., 2021, IL13Rα1 prevents a castration-resistant phenotype of prostate cancer by targeting hexokinase 2 for ubiquitin-mediated degradation, Cancer Biology & Medicine, 19: 1008-1028.
https://doi.org/10.20892/j.issn.2095-3941.2020.0583
PMID: 34652890 PMCID: PMC9334759
Hankey W., Chen Z., and Wang Q., 2020, Shaping chromatin states in prostate cancer by pioneer transcription factors, Cancer Research, 80(11): 2427-2436.
https://doi.org/10.1158/0008-5472.CAN-19-3447
PMID: 32094298 PMCID: PMC7299826
Heise M., Jarzemski P., Bąk A., Junkiert-Czarnecka A., Pilarska-Deltow M., and Haus O., 2019, G84E germline mutation in HOXB13 gene is associated with increased prostate cancer risk in polish men, Polish Journal of Pathology, 70(2): 127-133.
https://doi.org/10.5114/pjp.2019.87103
PMID: 31556563
Johng D., Haffner M.C., Mooney S.M., Esopi D.M., Ewing C.M., Chen S., and Isaacs W.B., 2016, Global analyses of HOXB13-regulated transcription reveal a potential link between HOXB13 G84E and prostate cancer risk, Cancer Research, 76: 2027-2027.
https://doi.org/10.1158/1538-7445.AM2016-2027
Kim E.H., Cao D., Mahajan N., Andriole G., and Mahajan K., 2020, ACK1–AR and AR–HOXB13 signaling axes: epigenetic regulation of lethal prostate cancers, NAR Cancer, 2: zcaa018.
https://doi.org/10.1093/narcan/zcaa018
PMID: 32885168 PMCID: PMC7454006
Kote-Jarai Z., Mikropoulos C., Leongamornlert D., Dadaev T., Tymrakiewicz M., Saunders E., Jones M., Jugurnauth-Little S., Govindasami K., and Guy M., 2015, Prevalence of the HOXB13 G84E germline mutation in british men and correlation with prostate cancer risk, tumour characteristics and clinical outcomes, Annals of Oncology, 26(4): 756-761.
https://doi.org/10.1093/annonc/mdv004
PMID: 25595936
Kurihara S., Matsui H., Ohtake N., Aoki M., Sekine Y., Arai S., Koike H., Suzuki K., and Miyazawa Y., 2022, Variants in HOXB13, G132E and F127C, are associated with prostate cancer risk in Japanese men, Cancer Diagnosis & Prognosis, 2(5): 542-548.
https://doi.org/10.21873/cdp.10139
PMID: 36060024 PMCID: PMC9425588
Liu C., Armstrong C., Ning S., Yang J.C., Lou W., Lombard A., Zhao J., Wu C., Yu A., Evans C., Tepper C., Li P.K., and Gao A., 2021, ARVib suppresses growth of advanced prostate cancer via inhibition of androgen receptor signaling, Oncogene, 40: 5379-5392.
https://doi.org/10.1038/s41388-021-01914-2
PMID: 34272475 PMCID: PMC8413131
Lotan T., Torres A.F.C., Zhang M., Tosoian J., Guedes L.B., Fedor H., Hicks J., Ewing C., Isaacs S., Johng D., De Marzo A.D., and Isaacs W., 2017, Somatic molecular subtyping of prostate tumors from HOXB13 G84E carriers, Oncotarget, 8: 22772-22782.
https://doi.org/10.18632/oncotarget.15196
Lu X., Fong K., Wang F., Gritsina G., Baca S., Berchuck J., Ross J., Corey E., Chandel N., Catalona W., Yang X.J., Freedman M., and Yu J., 2021, HOXB13 suppresses de novo lipogenesis through HDAC3-mediated epigenetic reprogramming, Nature Genetics, 54(5): 670-683.
https://doi.org/10.1038/s41588-022-01045-8.
PMID: 35468964 PMCID: PMC9117466
Maia S., Cardoso M., Pinto P., Pinheiro M., Santos C., Peixoto A., Bento M., Oliveira J., Henrique R., Jerónimo C., and Teixeira M., 2015, Identification of two novel HOXB13 germline mutations in portuguese prostate cancer patients, PLoS ONE, 10(7): e0132728.
https://doi.org/10.1371/journal.pone.0132728
PMID: 26176944 PMCID: PMC4503425
Misawa A., Kondo Y., Takei H., and Takizawa T., 2021, Long noncoding RNA HOXA11-AS and transcription factor HOXB13 modulate the expression of bone metastasis-related genes in prostate cancer, Genes, 12(2): 182.
https://doi.org/10.3390/genes12020182
Nerlakanti N., Yao J., Nguyen D., Patel A.K., Eroshkin A., Lawrence H., Ayaz M., Kuenzi B., Agarwal N., Chen Y., Gunawan S., Karim R., Berndt N., Puskás J., Magliocco A., Coppola D., Dhillon J., Zhang J., Shymalagovindarajan S., Rix U., Kim Y., Perera R., Lawrence N., Schonbrunn E., and Mahajan K., 2018, Targeting the BRD4-HOXB13 coregulated transcriptional networks with bromodomain-kinase inhibitors to suppress metastatic castration-resistant prostate cancer, Molecular Cancer Therapeutics, 17(12): 2796-2810.
https://doi.org/10.1158/1535-7163.MCT-18-0602
PMID: 30242092 PMCID: PMC6528782
Nguyen D., Yang W., Renganathan A., Weimholt C., Angappulige D.H., Nguyen T., Patel A.K., Agarwal N., Teer J., Coppola D., Zhang J., Perera R., and Mahajan K., 2022, Acetylated HOXB13 regulated super enhancer genes define therapeutic vulnerabilities of castration-resistant prostate cancer, Clinical Cancer Research, 28(18): 4131-4145.
https://doi.org/10.1158/1078-0432.CCR-21-3603
PMID: 35849143 PMCID: PMC9481728
Park C., Shin S.J., Cho Y., Joo J.W., and Cho N., 2019, HOXB13 expression in ductal type adenocarcinoma of prostate: clinicopathologic characteristics and its utility as potential diagnostic marker, Scientific Reports, 9(1): 20205.
https://doi.org/10.1038/s41598-019-56657-8
PMID: 31882852 PMCID: PMC6934792
Patel R.A., Sayar E., Coleman I.M., Roudier M.P., Hanratty B., Low J., Jaiswal N., Ajkunic A., Dumpit R., and Ercan C., 2023, Characterization of HOXB13 expression patterns in localized and metastatic castration‐resistant prostate cancer, The Journal of Pathology, 262.
https://doi.org/10.1002/path.6216
PMID: 37850574 PMCID: PMC10871027
Schnoeller T., Luedeke M., Rinckleb A., Stanford J., FitzGerald L., Schleutker J., Wahlfors T., Eeles R., Kote-Jarai Z., Weikert S., Krause H., Herkommer K., and Maier C., 2015, Association of the prostate cancer risk mutation G84E in HOXB13 with the subtype of ETS fusion negative adenocarcinoma with early age of diagnosis, Journal of Clinical Oncology, 33(15): 5021.
https://doi.org/10.1200/JCO.2015.33.15_SUPPL.5021
Sharp A., Coleman I.M., Yuan W., Sprenger C., Dolling D., Rodrigues D., Russo J.W., Figueiredo I., Bertan C., Seed G., Riisnaes R., Uo T., Neeb A., Welti J., Morrissey C., Luo J., Nelson P., Balk S., True L., de Bono J.D., and Plymate S., 2018, Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer, Journal of Clinical Investigation, 129(1): 192-208.
https://doi.org/10.1172/JCI122819
PMID: 30334814 PMCID: PMC6307949
Sipeky C., Gao P., Zhang Q., Wang L., Ettala O., Talala K., Tammela T., Auvinen A., Wiklund F., and Schleutker J., 2018, Synergistic interaction of HOXB13 and CIP2A predisposes to aggressive prostate cancer, Clinical Cancer Research, 24: 6265-6276.
https://doi.org/10.1158/1078-0432.CCR-18-0444
Song R.S., Sun K., Wang Y.X., Liu S.K., and Bu Y.Y., 2024, Synthetic microbial communities: redesigning genetic pathways for enhanced functional synergy, Molecular Microbiology Research, 14(1): 39-48.
https://doi.org/10.5376/mmr.2024.14.0005
Tilki D., Schaeffer E., and Evans C., 2016, Understanding Mechanisms of Resistance in Metastatic Castration-resistant Prostate Cancer: The Role of the Androgen Receptor, European Urology Focus, 2(5): 499-505.
https://doi.org/10.1016/j.euf.2016.11.013
PMID: 28723515
Wang T., 2024, The application prospects of immunomodulators in cancer treatment, Cancer Genetics and Epigenetics, 12(1): 1-7.
https://doi.org/10.5376/cge.2024.12.0001
Weiner A., Faisal F.A., Davicioni E., Karnes R.J., Vander Griend D.J., Lotan T., and Schaeffer E., 2020, Somatic HOXB13 expression correlates with metastatic progression in men with localized prostate cancer following radical prostatectomy, European Urology Oncology, 4(6): 955-962.
https://doi.org/10.1016/j.euo.2020.05.001
PMID: 32540218 PMCID: PMC7736205
Yao J., Chen Y., Nguyen D., Thompson Z., Eroshkin A., Nerlakanti N., Patel A.K., Agarwal N., Teer J., Dhillon J., Coppola D., Zhang J., Perera R., and Mahajan K., 2019, The Homeobox gene, HOXB13, regulates a mitotic protein-kinase interaction network in metastatic prostate cancers, Scientific Reports, 9: Article 46064.
https://doi.org/10.1038/s41598-019-46064-4
PMID: 31273254 PMCID: PMC6609629
Zabalza C.V., Adam M., Burdelski C., Wilczak W., Wittmer C., Kraft S., Krech T., Steurer S., Koop C., and Hube-Magg C., 2015, HOXB13 overexpression is an independent predictor of early PSA recurrence in prostate cancer treated by radical prostatectomy, Oncotarget, 6: 12822-12834.
https://doi.org/10.18632/oncotarget.3431
PMID: 25825985 PMCID: PMC4494977
(3).png)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Jianmin Wang
Related articles
. HOXB13
. Prostate cancer
. Genetic variants
. Androgen receptor signaling
. Personalized treatment
Tools
. Post a comment
.png)